Note
Click here to download the full example code
Compute transition frequency from custom clusters¶
This examples shows how to define two custom clusters of sensors and how to compute the corresponding theta-to-alpha transition frequency.
import mne
from transfreq import compute_transfreq_manual
from transfreq.viz import (plot_psds, plot_coefficients, plot_channels,
plot_clusters, plot_transfreq)
from transfreq.utils import read_sample_datapath
import os.path as op
import numpy as np
import matplotlib.pyplot as plt
# Define path to the data
subj = 'transfreq_sample'
data_folder = read_sample_datapath()
f_name = op.join(data_folder, '{}_resting.fif'.format(subj))
# Load resting state data
raw = mne.io.read_raw_fif(f_name)
# List of good channels
tmp_idx = mne.pick_types(raw.info, eeg=True, exclude='bads')
ch_names = [raw.ch_names[ch_idx] for ch_idx in tmp_idx]
# Compute power spectra
n_fft = 512*2
bandwidth = 1
fmin = 2
fmax = 30
sfreq = raw.info['sfreq']
n_per_seg = int(sfreq*2)
psds, freqs = mne.time_frequency.psd_multitaper(raw, fmin=fmin, fmax=fmax,
bandwidth=bandwidth)
# Read channel positions
ch_locs = np.zeros((psds.shape[0], 3))
for ii in range(psds.shape[0]):
ch_locs[ii, :] = raw.info['chs'][ii]['loc'][:3]
Out:
Opening raw data file /home/sara/Documenti/San_Martino_Alzhaimer/transition_frequency/transfreq/data/transfreq_sample_resting.fif...
/home/sara/Documenti/San_Martino_Alzhaimer/transition_frequency/examples/plot_manual_analysis.py:25: RuntimeWarning: This filename (/home/sara/Documenti/San_Martino_Alzhaimer/transition_frequency/transfreq/data/transfreq_sample_resting.fif) does not conform to MNE naming conventions. All raw files should end with raw.fif, raw_sss.fif, raw_tsss.fif, _meg.fif, _eeg.fif, _ieeg.fif, raw.fif.gz, raw_sss.fif.gz, raw_tsss.fif.gz, _meg.fif.gz, _eeg.fif.gz or _ieeg.fif.gz
raw = mne.io.read_raw_fif(f_name)
Range : 0 ... 29558 = 0.000 ... 59.116 secs
Ready.
Using multitaper spectrum estimation with 57 DPSS windows
Plot power spectrum to visually chose theta and alpha ranges
plot_psds(psds, freqs, average=True)
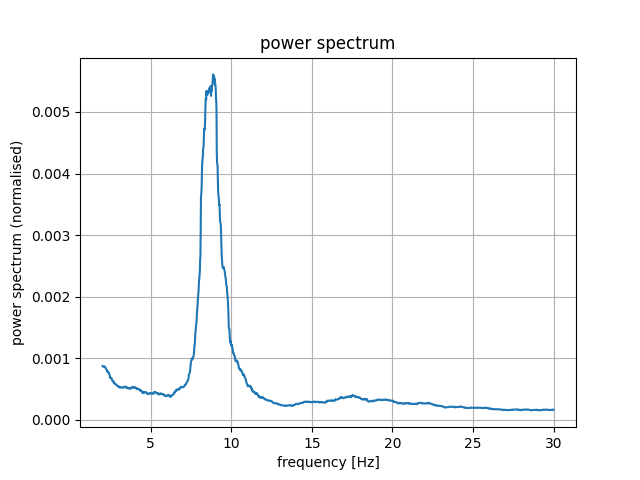
Out:
<Figure size 640x480 with 1 Axes>
set theta and alpha ranges
alpha_range = [8, 9.5]
theta_range = [6.5, 7]
Plot alpha and theta coefficients. In the 1D visualisation the ratio between alpha and theta coefficients is plotted. In the 2d visualisation a scatter-plot of the alpha and theta coefficients is shown.
fig = plt.figure()
gs = fig.add_gridspec(2, 2)
ax1 = fig.add_subplot(gs[0, :])
plot_coefficients(psds, freqs, ch_names=ch_names, alpha_range=alpha_range,
theta_range=theta_range, mode='1d', ax=ax1, order='sorted')
ax2 = fig.add_subplot(gs[1, 0])
plot_coefficients(psds, freqs, ch_names=ch_names, alpha_range=alpha_range,
theta_range=theta_range, mode='2d', ax=ax2)
# Plot the corresponding averaged power spectra.
ax3 = fig.add_subplot(gs[1, 1])
plot_psds(psds, freqs, average=True, ax=ax3)
fig.tight_layout()
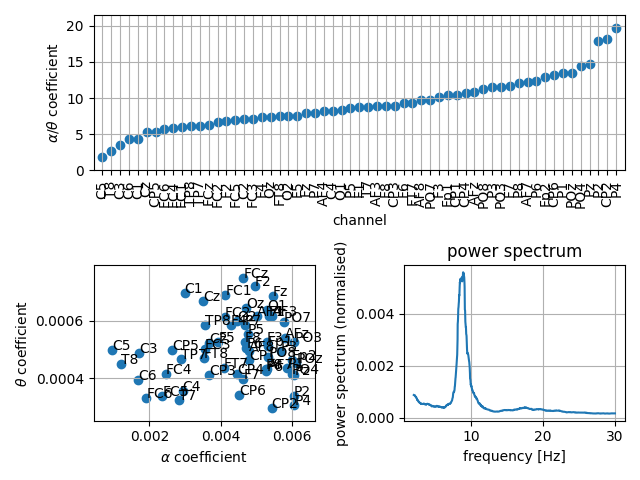
Chose channels from the figure to manually define the clusters and compute the corresponding transition frequency
# First definition by looking at the 1D plot
theta_chs_1d = ['C5', 'C3', 'T8', 'C1', 'C6']
alpha_chs_1d = ['P4', 'P2', 'CP2']
# Second definition by looking at the 2D plot
theta_chs_2d = ['C5', 'C3', 'T8']
alpha_chs_2d = ['P4', 'P2', 'Pz', 'CP2', 'POz', 'Fp2', 'PO4', 'P1']
tfbox_1d = compute_transfreq_manual(psds, freqs, theta_chs_1d, alpha_chs_1d,
ch_names=ch_names, theta_range=theta_range,
alpha_range=alpha_range, method='my_method_1d')
tfbox_2d = compute_transfreq_manual(psds, freqs, theta_chs_2d, alpha_chs_2d,
ch_names=ch_names, theta_range=theta_range,
alpha_range=alpha_range, method='my_method_2d')
Plot results obtained with the first definition of the clusters (1D)
fig = plt.figure(constrained_layout=True, figsize=(15, 10))
subfigs = fig.subfigures(2, 2, wspace=0.1)
plot_channels(tfbox_1d, ch_locs, mode='1d', subfig=subfigs[0,0])
ax1 = subfigs[0,1].subplots(1, 1)
plot_clusters(tfbox_1d, mode='1d', order='sorted', ax=ax1)
ax2 = subfigs[1,0].subplots(1, 1)
plot_transfreq(psds, freqs, tfbox_1d, ax=ax2)
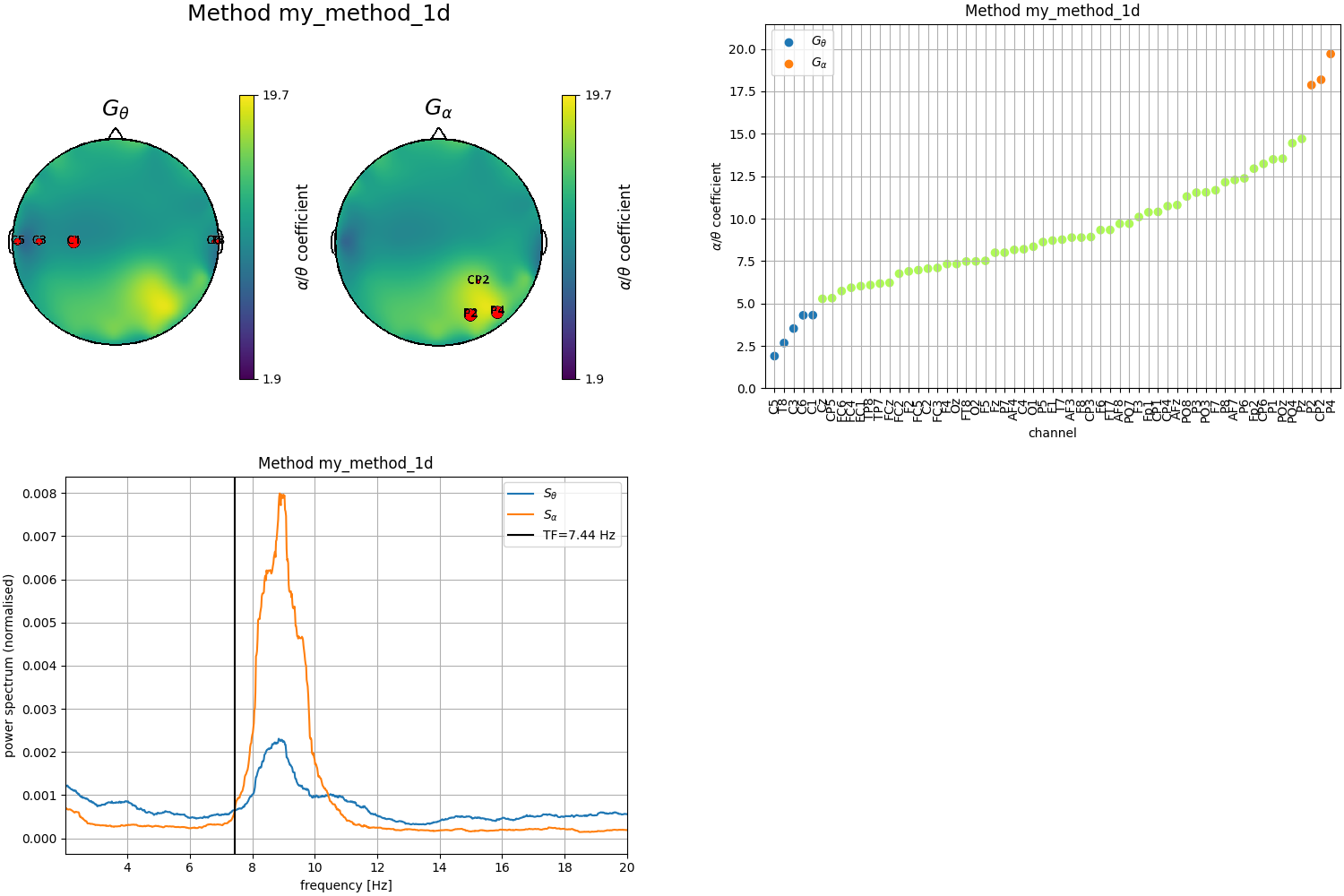
Out:
<matplotlib.figure.SubFigure object at 0x7fafa498d310>
Plot results obtained with the second definition of the clusters (2D)
fig = plt.figure(constrained_layout=True, figsize=(15, 10))
subfigs = fig.subfigures(2, 2, wspace=0.07)
plot_channels(tfbox_2d, ch_locs, mode='2d', subfig=subfigs[0,0])
ax1 = subfigs[0,1].subplots(1, 1)
plot_clusters(tfbox_2d, mode='2d', order='sorted', ax=ax1)
ax2 = subfigs[1,0].subplots(1, 1)
plot_transfreq(psds, freqs, tfbox_2d, ax=ax2)
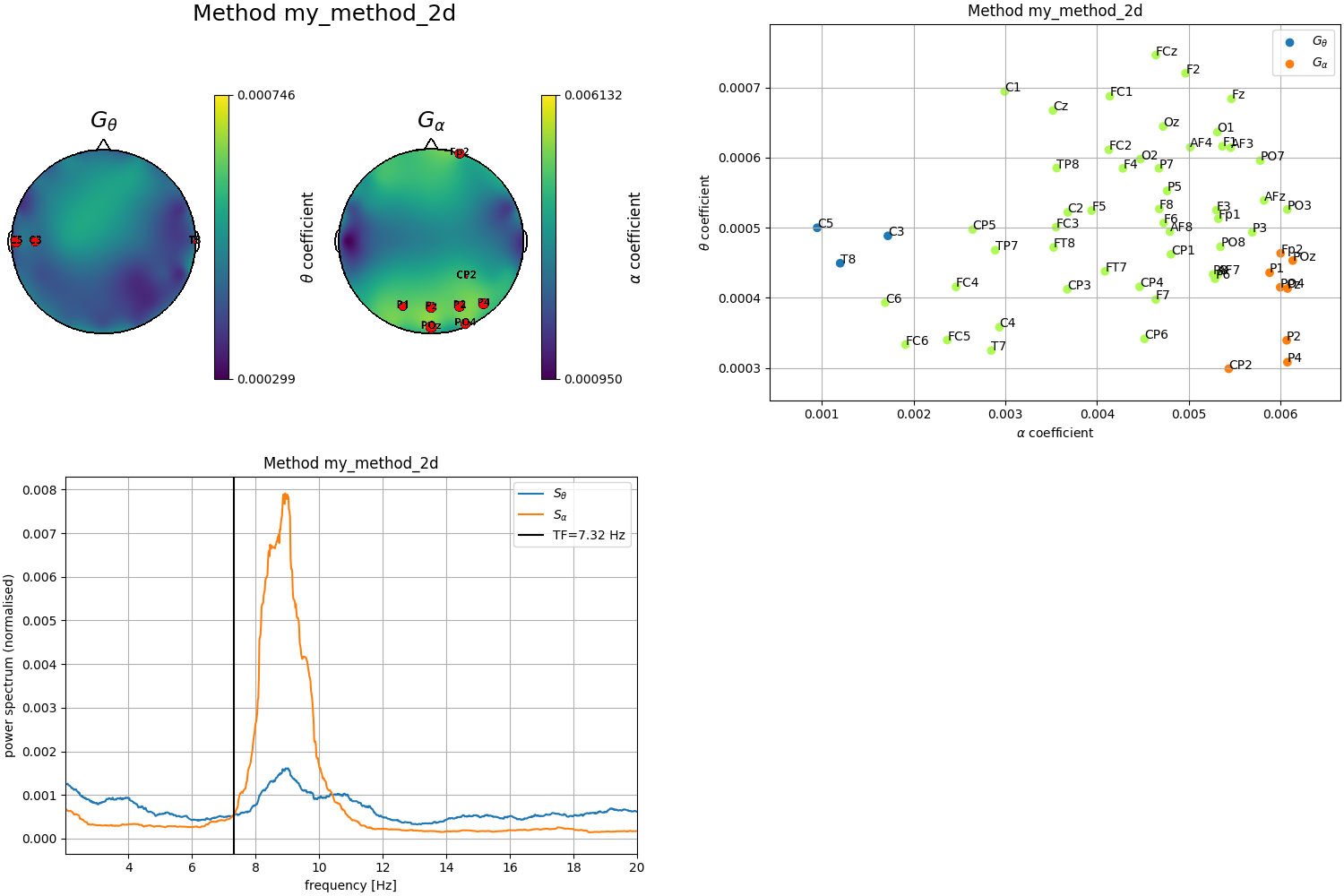
Out:
/home/sara/Documenti/San_Martino_Alzhaimer/transition_frequency/transfreq/viz.py:40: UserWarning: order can only be 'standard' if mode is '2d' or mode is None and method in tfbox is 3 or 4. Automatically switched to 'standard'
"order can only be 'standard' if mode is '2d' or mode is None and method in tfbox is 3 or 4. Automatically switched to 'standard'")
<matplotlib.figure.SubFigure object at 0x7faf96f745d0>
Total running time of the script: ( 0 minutes 26.846 seconds)